Integrating Theory, Computation, and Experiments to Robustly Design Complex Protein-based Nanomaterials

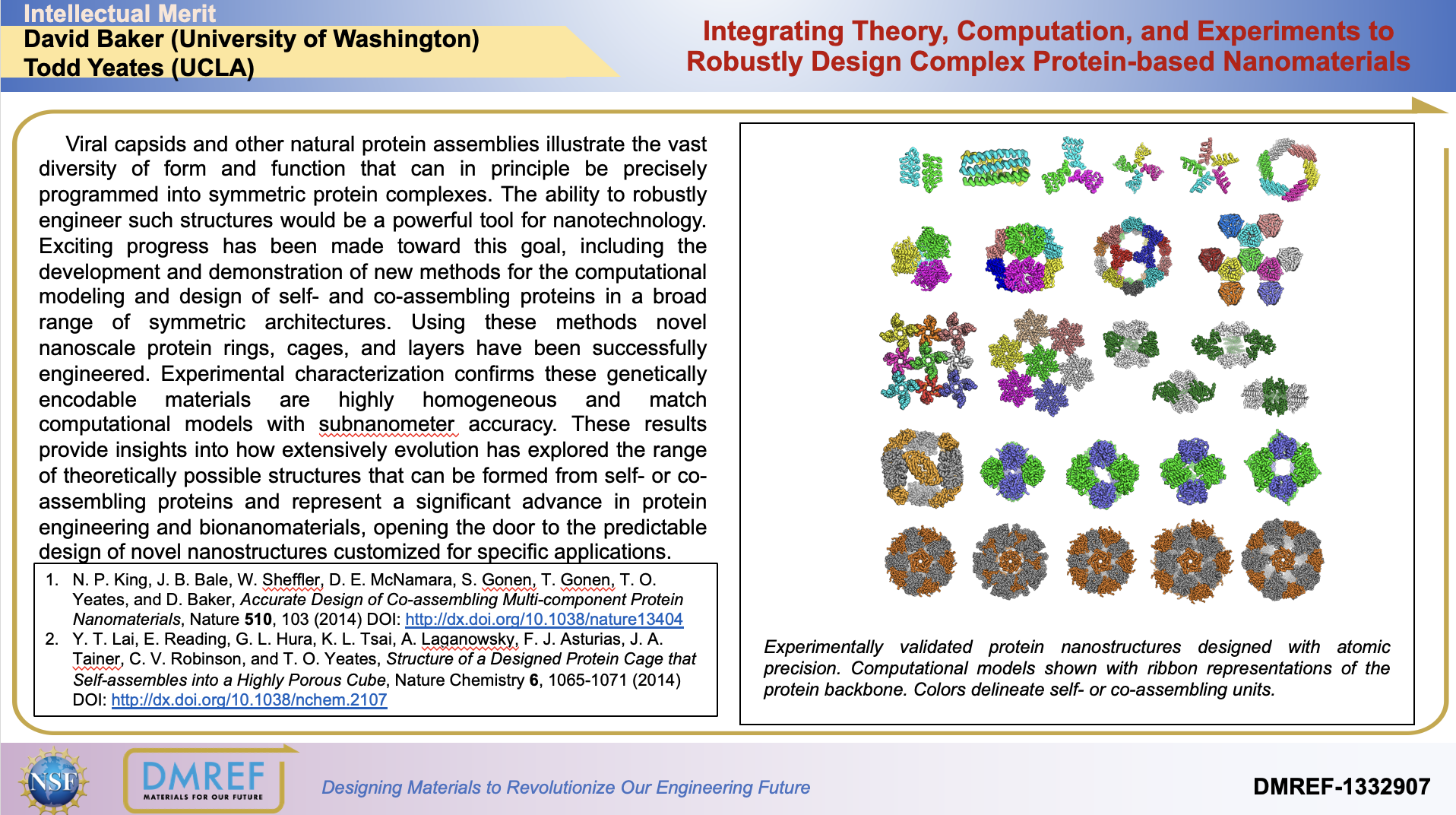
Viral capsids and other natural protein assemblies illustrate the vast diversity of form and function that can in principle be precisely programmed into symmetric protein complexes. The ability to robustly engineer such structures would be a powerful tool for nanotechnology. Exciting progress has been made toward this goal, including the development and demonstration of new methods for the computational modeling and design of self- and co-assembling proteins in a broad range of symmetric architectures.
Using these methods novel nanoscale protein rings, cages, and layers have been successfully engineered. Experimental characterization confirms these genetically encodable materials are highly homogeneous and match computational models with subnanometer accuracy. These results provide insights into how extensively evolution has explored the range of theoretically possible structures that can be formed from self- or co-assembling proteins and represent a significant advance in protein engineering and bionanomaterials, opening the door to the predictable design of novel nanostructures customized for specific applications.